Reproducible Research
Posted on Wed 01 January 2020 in research
One of the key principles of proper scientific procedure is the act of repeating an experiment or analysis and being able to reach similar conclusions. Published research based on computational analysis, e.g. bioinformatics or computational biology, have often suffered from incomplete method descriptions (e.g. list of used software versions); unavailable raw data; and incomplete, undocumented and/or unavailable code. This essentially prevents any possibility of attempting to reproduce the results of such studies. The term "reproducible research" has been used to describe the idea that a scientific publication based on computational analysis should be distributed along with all the raw data and metadata used in the study, all the code and/or computational notebooks needed to produce results from the raw data, and the computational environment or a complete description thereof.
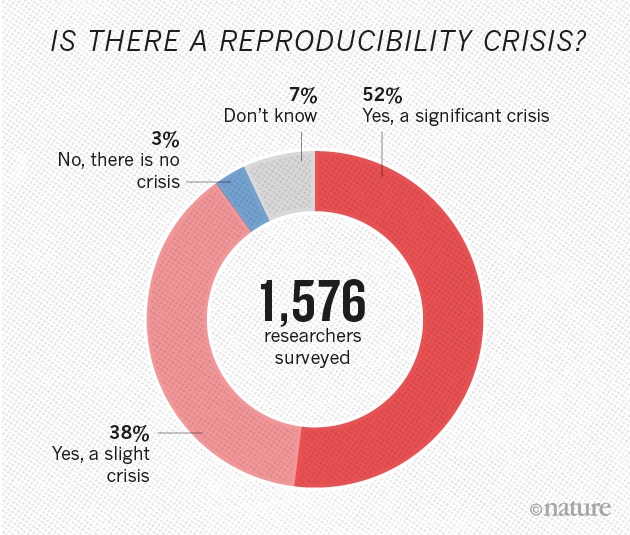
From: M. Baker, 1,500 scientists lift the lid on reproducibility, Nature, vol. 533, no. 7604, pp. 452–454, May 2016, https://doi.org/10.1038/533452a.
Reproducible research not only leads to proper scientific conduct but also provides other researchers the access to build upon previous work. Most importantly, the person setting up a reproducible research project will quickly realize the immediate personal benefits: an organized and structured way of working. The person that most often has to reproduce your own analysis is your future self!
All our work is driven by fully reproducible framework. More recently, we have been actively looking at the reproducibility of published work and how to lower the entrance barrier of publication readers. We argue it is insufficient, in most cases, to only publish software leading to results if original data remains inaccessible. Reproducibility should imply in the following characteristics: repeatability, share-ability, extensibility and stability, which is not guaranteed by most published material to date (Anjos et al., 2017). We propose a software suite called Bob that possesses these characteristics, demonstrating its flexibility to various tasks including Medical Image Segmentation, Biometric Person Recognition, Presentation Attack Detection, and Remote Photoplethysmography.
From another perspective, there are legitimate cases in which raw data leading to research conclusions cannot be published. Furthermore, in a growing number of use-cases, the availability of both software does not translate to an accessible reproducibility scenario. To bridge this gap, we built an open platform for research called BEAT (Anjos et al., 2017) in computational sciences related to pattern recognition and machine learning, to help on the development, reproducibility and certification of results obtained in the field.
Bob and BEAT are still active and support past and future work at Idiap and beyond. We also conduct lectures to both master and graduate students about reproducibility in data science.
Bibliography
André Anjos, Laurent El Shafey, and Sébastien Marcel. Beat: an open-science web platform. In Thirty-fourth International Conference on Machine Learning. August 2017. URL: https://publications.idiap.ch/index.php/publications/show/3665. ↩
André Anjos, Manuel Günther, Tiago de Freitas Pereira, Pavel Korshunov, Amir Mohammadi, and Sébastien Marcel. Continuously reproducing toolchains in pattern recognition and machine learning experiments. In Thirty-fourth International Conference on Machine Learning. August 2017. URL: https://publications.idiap.ch/index.php/publications/show/3666. ↩