Reproducible Research and Computing Platforms
Posted on Wed 25 September 2024 in research
One of the key principles of proper scientific procedure is the act of repeating an experiment or analysis and being able to reach similar conclusions. Published research based on computational analysis, e.g. bioinformatics or computational biology, have often suffered from incomplete method descriptions (e.g. list of used software versions); unavailable raw data; and incomplete, undocumented and/or unavailable code. This essentially prevents any possibility of attempting to reproduce the results of such studies. The term "reproducible research" has been used to describe the idea that a scientific publication based on computational analysis should be distributed along with all the raw data and metadata used in the study, all the code and/or computational notebooks needed to produce results from the raw data, and the computational environment or a complete description thereof.
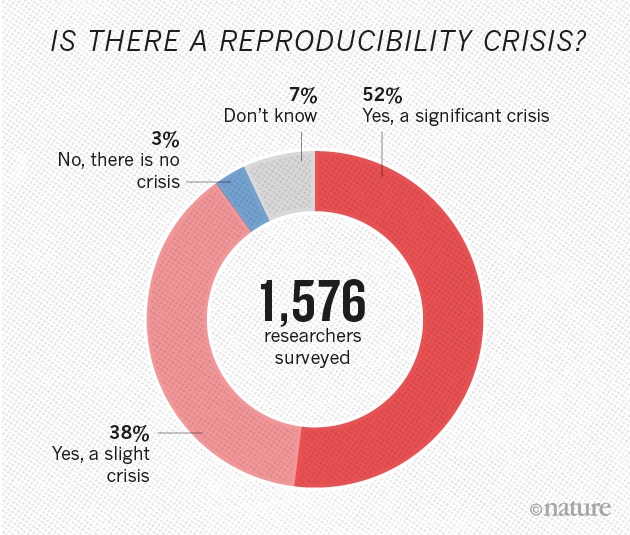
From: M. Baker, 1,500 scientists lift the lid on reproducibility, Nature, vol. 533, no. 7604, pp. 452–454, May 2016, https://doi.org/10.1038/533452a.
Reproducible research not only leads to proper scientific conduct but also provides other researchers the access to build upon previous work. Most importantly, the person setting up a reproducible research project will quickly realize the immediate personal benefits: an organized and structured way of working. The person that most often has to reproduce your own analysis is your future self!
All our work is driven by a fully reproducible framework. As such, I'm always active in evaluating the reproducibility of published research and seeking ways to lower the barriers for contributions. In most cases, it's insufficient simply to publish software that leads to results without making original data accessible. A truly reproducible workflow should possess characteristics such as repeatability, shareability, extensibility, and stability – features that are not consistently guaranteed by most published work [1].
We typically propose software suites (e.g., mednet, Bob) that embody these desirable traits. The diverse applications of these suites demonstrate the flexibility of our approach to various tasks, including medical image classification, segmentation, and object detection; biometric person recognition; presentation attack detection; and remote photoplethysmography.
From another perspective, there are legitimate cases in which raw data leading to research conclusions cannot be published. Furthermore, in a growing number of use-cases, the availability of both software does not translate into an accessible reproducibility scenario. To bridge this gap, we built a prototype open platform for research called BEAT [2]. This platform provided a computing infrastructure for open science, proposing solutions for open access to scientific information, sharing, and re-use – including data and source code while protecting privacy and confidentiality. After nearly 10 years of usage and development, the BEAT platform is now retired.
Our recently granted project, CollabCloud, further investigates how web-accessible computing platforms can enhance reproducibility in data sciences, while also increasing collaboration potential for computing infrastructures. Researchers conducting Artificial Intelligence (AI) breakthrough research often face significant barriers due to limited access to resources, including well-tuned systems, large datasets, and computing power. This slows down complementary research as teams spend a substantial amount of time setting up and transferring systems between institutions. To address this issue, the Idiap Research Institute is developing "CollabCloud", a cloud-based research infrastructure designed to boost collaborative work in AI research. The platform aims to provide easy exportability of research workflows, controlled access to shared storage and computing power, and the ability for both computational and non-computational experts to explore AI models together. CollabCloud uses the Renku platform from the Swiss Data Science Center.
Bibliography
[1] André Anjos, Manuel Günther, Tiago de Freitas Pereira, Pavel Korshunov, Amir Mohammadi, and Sébastien Marcel. Continuously reproducing toolchains in pattern recognition and machine learning experiments. In Thirty-fourth International Conference on Machine Learning. August 2017. URL: https://publications.idiap.ch/index.php/publications/show/3666.
@inproceedings{icml-2017-2,
author = "Anjos, André and Günther, Manuel and de Freitas Pereira, Tiago and Korshunov, Pavel and Mohammadi, Amir and Marcel, Sébastien",
month = "August",
title = "Continuously Reproducing Toolchains in Pattern Recognition and Machine Learning Experiments",
booktitle = "Thirty-fourth International Conference on Machine Learning",
year = "2017",
location = "Sidney, Australia",
url = "https://publications.idiap.ch/index.php/publications/show/3666",
pdf = "https://www.idiap.ch/\textasciitilde aanjos/papers/icml-2017-2.pdf",
poster = "https://www.idiap.ch/\textasciitilde aanjos/posters/icml-2017-2.pdf",
abstract = "Pattern recognition and machine learning research work often contains experimental results on real-world data, which corroborates hypotheses and provides a canvas for the development and comparison of new ideas. Results, in this context, are typically summarized as a set of tables and figures, allowing the comparison of various methods, highlighting the advantages of the proposed ideas. Unfortunately, result reproducibility is often an overlooked feature of original research publications, competitions, or benchmark evaluations. The main reason for such a gap is the complexity on the development of software associated with these reports. Software frameworks are difficult to install, maintain, and distribute, while scientific experiments often consist of many steps and parameters that are difficult to report. The increasingly rising complexity of research challenges make it even more difficult to reproduce experiments and results. In this paper, we emphasize that a reproducible research work should be repeatable, shareable, extensible, and stable, and discuss important lessons we learned in creating, distributing, and maintaining software and data for reproducible research in pattern recognition and machine learning. We focus on a specific use-case of face recognition and describe in details how we can make the recognition experiments reproducible in practice."
}
[2] André Anjos, Laurent El Shafey, and Sébastien Marcel. Beat: an open-science web platform. In Thirty-fourth International Conference on Machine Learning. August 2017. URL: https://publications.idiap.ch/index.php/publications/show/3665.
@inproceedings{icml-2017-1,
author = "Anjos, André and El Shafey, Laurent and Marcel, Sébastien",
month = "August",
title = "BEAT: An Open-Science Web Platform",
booktitle = "Thirty-fourth International Conference on Machine Learning",
year = "2017",
location = "Sydney, Australia",
url = "https://publications.idiap.ch/index.php/publications/show/3665",
pdf = "https://www.idiap.ch/\textasciitilde aanjos/papers/icml-2017-1.pdf",
poster = "https://www.idiap.ch/\textasciitilde aanjos/posters/icml-2017-1.pdf",
abstract = "With the increased interest in computational sciences, machine learning (ML), pattern recognition (PR) and big data, governmental agencies, academia and manufacturers are overwhelmed by the constant influx of new algorithms and techniques promising improved performance, generalization and robustness. Sadly, result reproducibility is often an overlooked feature accompanying original research publications, competitions and benchmark evaluations. The main reasons behind such a gap arise from natural complications in research and development in this area: the distribution of data may be a sensitive issue; software frameworks are difficult to install and maintain; Test protocols may involve a potentially large set of intricate steps which are difficult to handle. To bridge this gap, we built an open platform for research in computational sciences related to pattern recognition and machine learning, to help on the development, reproducibility and certification of results obtained in the field. By making use of such a system, academic, governmental or industrial organizations enable users to easily and socially develop processing toolchains, re-use data, algorithms, workflows and compare results from distinct algorithms and/or parameterizations with minimal effort. This article presents such a platform and discusses some of its key features, uses and limitations. We overview a currently operational prototype and provide design insights."
}